<html>
<script src="http://www.google-analytics.com/urchin.js"
type="text/javascript">
</script>
<script type="text/javascript">
_uacct = "UA-564843-2";
urchinTracker();
</script>
</html>
<html><style type='text/css'>
A:link {
text-decoration: underline;
font-size: 10pt;
font-style: normal;
font-family: arial, trebuchet ms;
color: #003399;
}
A:visited {
text-decoration: underline;
font-size: 10pt;
font-style: normal;
font-family: arial, trebuchet ms;
color: #003399;
}
A:active {
text-decoration: none;
background-color: #99ccff;
color: #660099;
}
A:hover {
text-decoration: none;
color: #ff0033;
}
.navbar{
padding: 0px;
width: "100%";
font-family: Arial;
font-size: 10pt;
}
</style></html>
|
|
Florian M. Freimoser, PhD
Institute of Plant Sciences
Biochemistry and Physiology of Plants
ETH Zurich, LFW D46.1
Universitätsstr. 2
CH-8092 Zurich
Switzerland
Tel: +41 +44 632 38 44
Fax: +41 +44 632 10 44
Please also visit our other web site!
|
The molecule we study is so plain
the inorganic polyphosphate chain
Our group studies a simple
molecule: inorganic
polyphosphate (poly P).
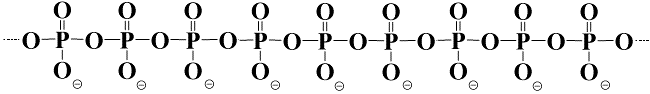
Poly P is a linear polymer that consists of a variable number of
phosphate residues
(from three to more than a thousand) that are
linked by energy-rich phosphoanhydride bonds. It occurs ubiquitously in all living cells and regulates many molecular and biological processes. Nevertheless, poly P is scarcely studied and little is known about poly P metabolism and its exact molecular functions; especially in eukaryotes. In our group we have developed methods to quantify poly P, to stain and localize poly P and to screen for poly P binding proteins. We are using these tools to investigate poly P metabolism and functions in fungi, plants and algae. However, at the moment our main efforts are devoted to the study of poly P metabolism in the yeast Saccharomyces cerevisiae and of poly P in fungal cell walls.
|